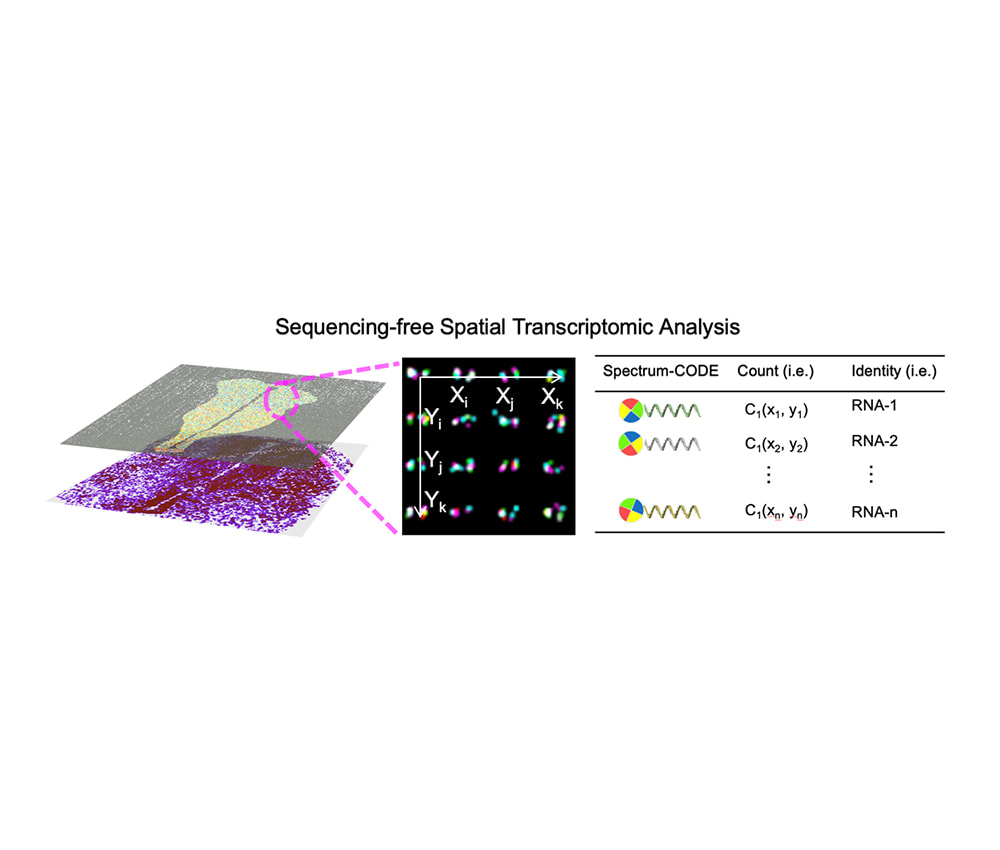
Tissue spatial transcriptomics
Leveraging our proprietary spatial omics biochip technology, gene capture is performed in situ on tissue sections at single-cell to subcellular resolution. RNA molecules are labeled using fluorescent in situ hybridization (FISH) and combined with barcode encoding to enable high-throughput gene profiling, providing high-resolution spatial gene expression data.
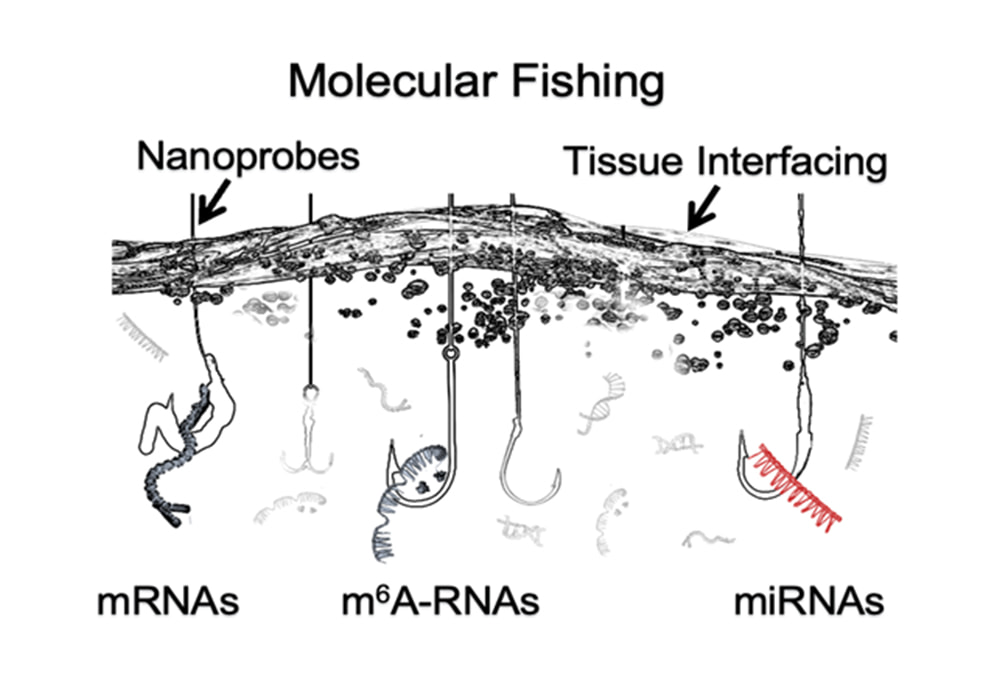
Technology
Chemically modified nano-needle arrays penetrate the cell membrane, specifically capturing RNA or molecular beacons through the modification layer to detect target RNA. Integrated spatial barcodes preserve the positional information of RNA molecules, enabling reconstruction of the spatial distribution of gene expression.
Scope
Fresh-frozen tissues, FFPE samples, live biopsy specimens, and whole-organ sections from animal models.
Advantages
Subcellular Precision
Nano-needle tips with diameters capable of penetrating the nuclear membrane directly capture mRNA, enabling gene expression analysis at the cytoplasmic level.
FFPE Sample Compatibility
Supports processing of clinically archived formalin-fixed, paraffin-embedded samples, making it suitable for large-scale retrospective studies.
Penetration of Complex Tissues
Diamond nano-needles can penetrate hard structures such as bone, expanding the applicability of spatial transcriptomics.
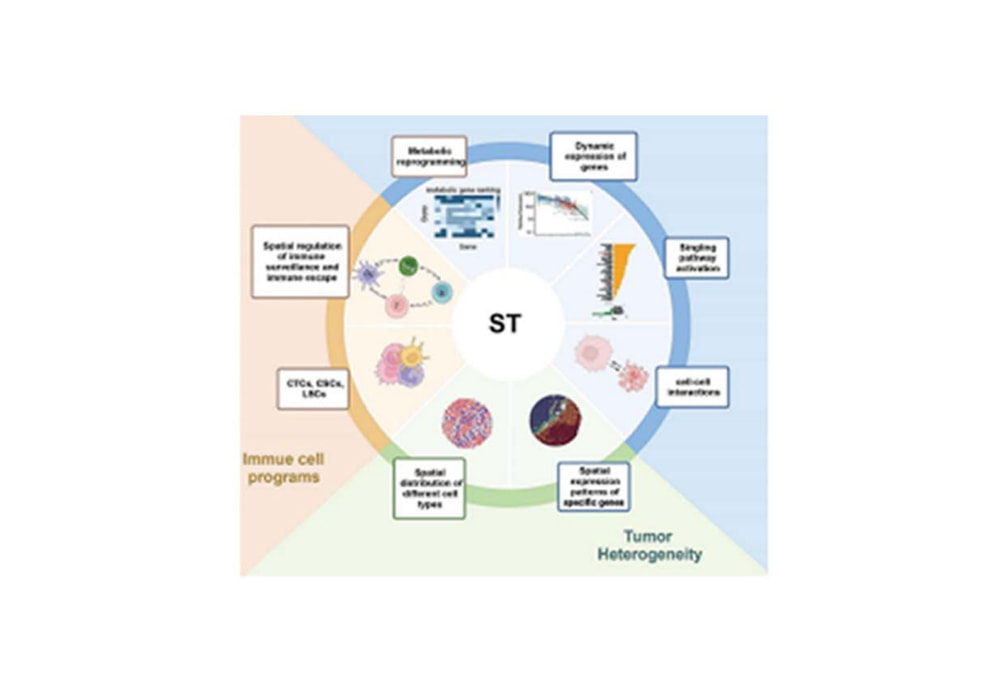
Application
Spatial transcriptomics enables multi-dimensional analysis of tumor heterogeneity, including immune regulation, metabolic reprogramming, and dynamic gene expression, covering aspects such as immune evasion and cell-cell interactions.
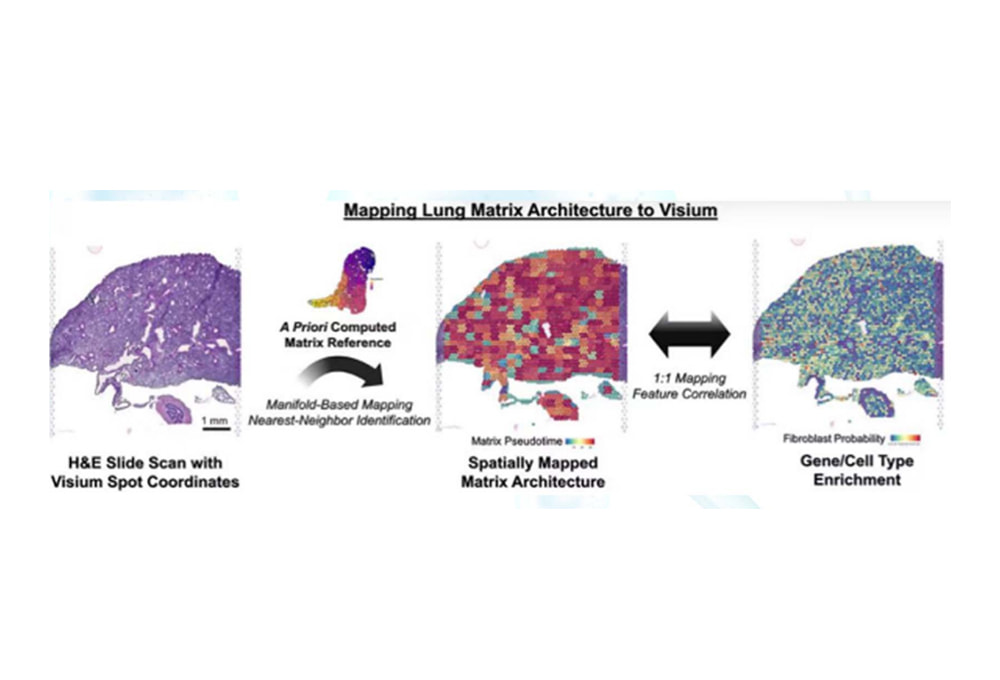
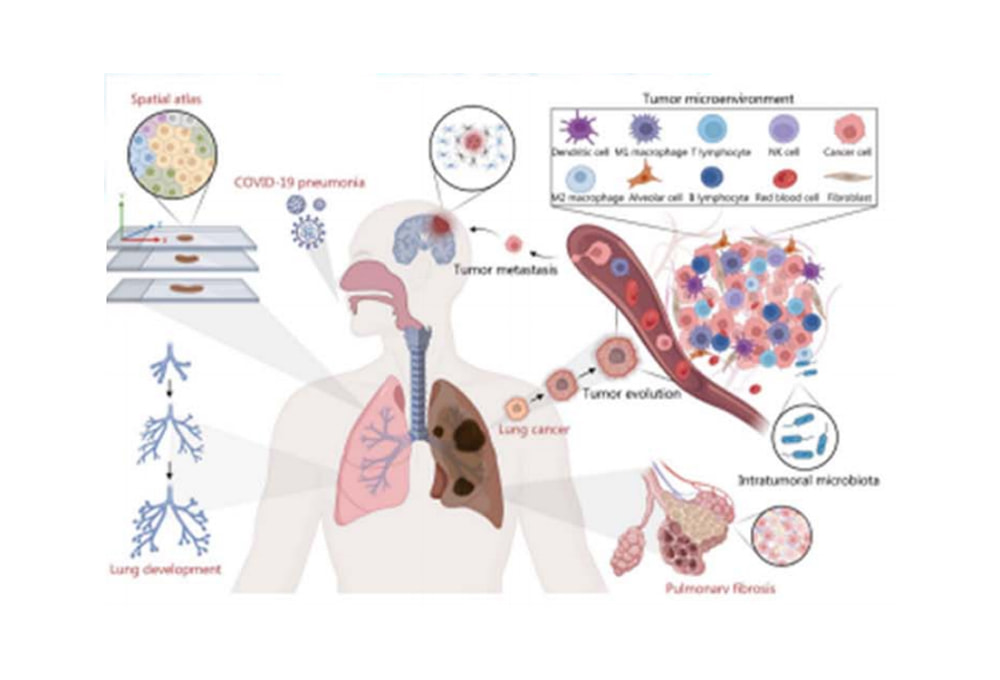
Using prior reference data of the stroma, manifold mapping and nearest-neighbor identification methods associate stromal structures with H&E-stained sections and Visium coordinates in a one-to-one manner. This allows spatial localization of stromal structures, analysis of fibroblast probabilities, and gene/cell-type enrichment, revealing the spatial molecular characteristics of lung tissue.
Spatial maps facilitate the study of spatial interactions between immune and cancer cells, tumor evolution, intratumoral microbiota, and pulmonary fibrosis mechanisms. Applications include COVID-19 pneumonia, tumor microenvironment analysis, and metastasis studies, covering lung development, lung cancer, and fibrosis, providing insights into tumor microenvironments and intratumoral microbiota.